Research
Microscopy: system & algorithm
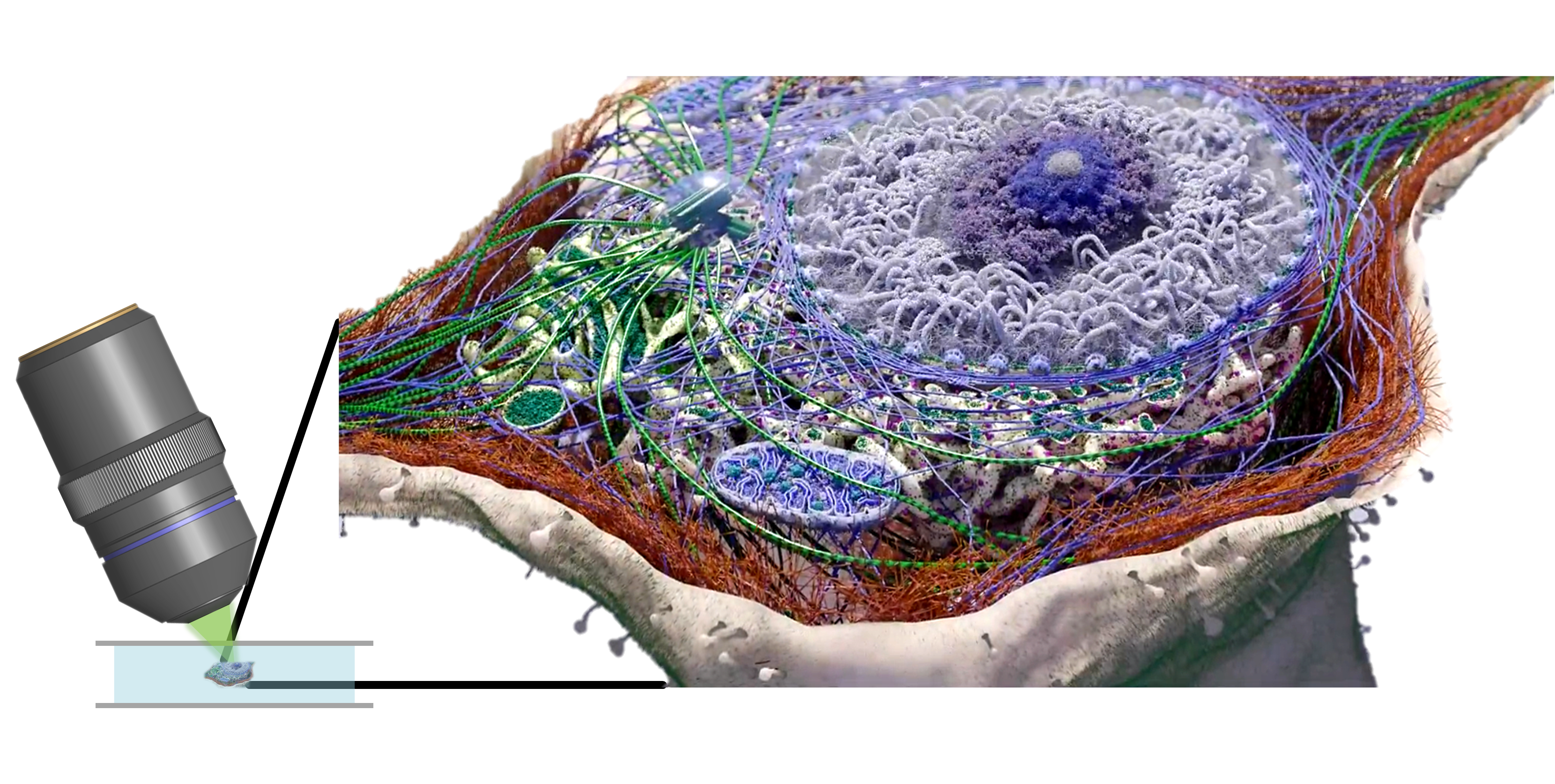
We are focusing on developing advanced optical microscopy for biomedical applications. He particularly is interested in new imaging physics, bottom-up opto-electronic system design, as well as harnessing / designing optimal algorithms / optics for every specific application scenario.
The techniques developed included the
(1) Sparse deconvolution algorithm for extending spatial resolution, contrast and SNR of fluorescence microscopes limited by the optics (applications: Sparse-SIM & Sparse SD-SIM);
(2) PANEL (pixel-level analysis of error locations) framework for quantitatively mapping reconstruction errors at super-resolution scale, which can assess both conventional and deep-learning methods;
(3) SACD (super-resolution method based on the auto-correlation with two-step deconvolution), a fully automatic super-resolution method that can realize a twofold 3D resolution improvement using only 20 frames without needing additional optical components.
Related technologies:
Label-free: Endoscopic/label-free-super-resolution/optical-diffraction-tomography microscopy;
Fluorescence: (miniaturized) SIM/SOFI/SMLM/light-field/multi-photon/confocal microscopy.
Machine learning: method & application
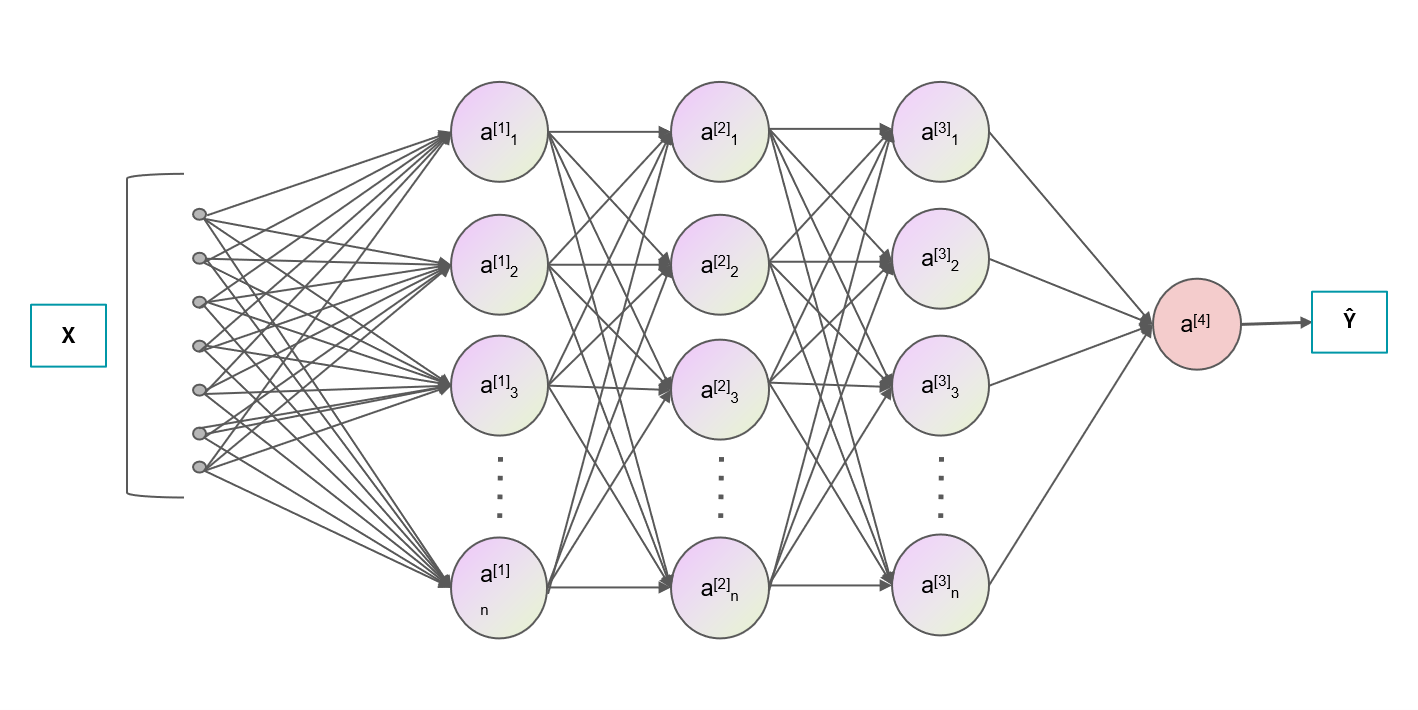
We are using the available prior knowledge to design specific algorithms to analyze and reconstruct the multi-dimensional bio/natural signals (non-deep model). On the other hand, taking advantage of priors from data, deep-learning has the fascinating character (e.g., automatic feature extraction) on solving the problems that can never be solved by conventional ways. He is also using deep-learning to reduce the photon / time / hardware budgets of microscopy, as well as employing (self-supervised, representation, Bayesian) deep-learning to speed up the data profiling pipeline of biologists.
The techniques developed included the
(1) SN2N (self-inspired Noise2Noise), an unsupervised learning-to-denoise engin, which is fully competitive with the supervised learning methods and overcomes the need for large dataset and clean ground-truth.
Related technologies:
Non-deep model: Deconvolution, denoise, phase retrieval, optical tracking, segmentation, and Bayesian inference.
Deep-learning: Cross-modality, low-level transformation, segmentation, visual-relationship.
Biomedical application & Smart bioimage analysis
We are devising high-level algorithms for bioimage analysis to help the biologists to automatically analyze large amounts of image data, reproducibly extract quantitative information from images, and quantify the dynamics, form, and structure of cells and organisms.
The techniques developed included the
(1) Smart palm-size optofluidic hematology analyzer for automated imaging-based leukocyte concentration detection.
(2) MIRD a highly flexible, compact, single-shot volumetric, high-resolution, endoscopic probe by employing precision-machined multiple micro-imaging devices (MIRD).
(3) Sparse confocal microscopy with single particle tracking for uncover that SARS-CoV-2 virus-like particles utilize filopodia to reach the entry site in two patterns, “surfing” and “grabbing”, which avoid the virus from randomly searching on the plasma membrane..

