Open-source
Sparse deconvolution
This repo. contains the update version of this software (Sparse deconvolutionn). It is an universal post-processing framework for fluorescent microscopy.
Basic features:
1. Denoise;
2. Deconvolution (3D resolution enhancement);
3. Upsample;
4. Remove background (epi to TIRF);
5. Remove artifacts;
SACD
1. SACD only needs 20 frames to achieve twofold lateral and axial resolution improvements.
2. SACD requires no additional optical components, and can offer a direct and flexible add-on super-resolution feature on any off-the-shelf commercialized microscopes or in-house customized systems.
3. Sparse deconvolution-assisted SACD (Sparse-SACD) allows fast 4D super-resolution live-cell imaging.
PANEL
rFRC: The rolling Fourier ring correlation (rFRC) is for quantitatively mapping the local image quality (effective resolution, data uncertainty). The lower effective resolution gives a higher probability to the error existence, and thus we can use it to represent the uncertainty revealing the error distribution.
PANEL: We also accompany our filtered rFRC with truncated RSM (resolution-scaled error map) as a full PANEL (pixel-level analysis of error locations) map, but this RSM is an optional feature that can be turn off as the wide-field reference being unavailable. PANEL is for biologists to qualitatively pinpoint regions with low reliability as a concise visualization.
SN2N
Our SN2N (self-inspired Noise2Noise) is fully competitive with the supervised learning methods and overcomes the need for large dataset and clean ground-truth. First, we create a self-supervised data generation strategy based on super-resolution images' spatial redundancy, using a diagonal resampling step followed by a Fourier interpolation for single-frame Noise2Noise. Second, we have taken a step further by ushering in a self-constrained learning process to enhance the performance and data-efficiency. Finally, we provide a Patch2Patch data augmentation (random patch transformations in multiple dimensions) to further improve the data efficiency.
Basic features:
1. Unsupervised learning to denoise;
2. Data augmentation.
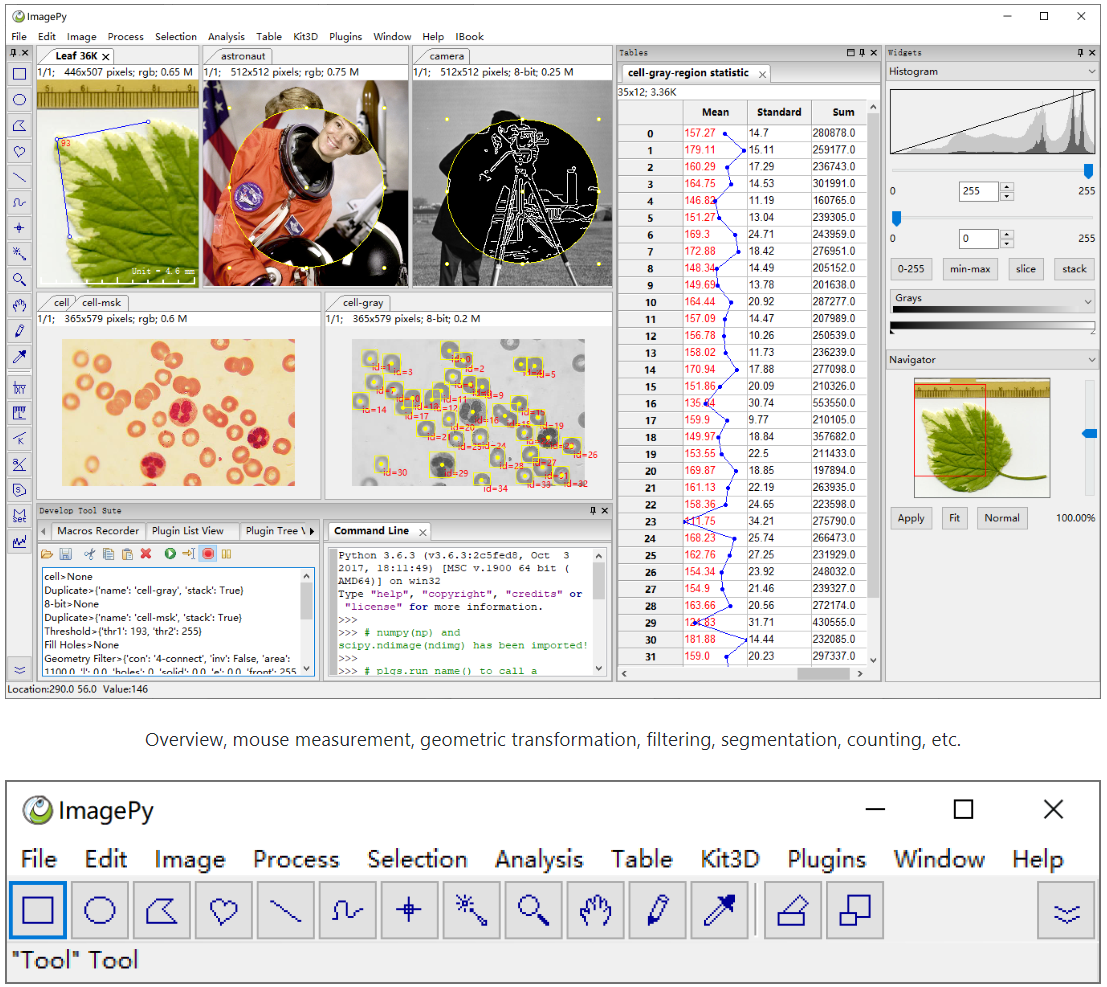
ImagePy
An open source image processing framework written in Python. Its UI interface, image data structure and table data structure are wxpython-based, Numpy-based and pandas-based respectively. Furthermore, it supports any plug-in based on Numpy and pandas, which can talk easily between scipy.ndimage, scikit-image, simpleitk, opencv and other image processing libraries.
A pythonic ImageJ:
1. Has a user-friendly interface
2. Can read/save a variety of image data formats
3. Supports ROI settings, drawing, measurement and other mouse operations
4. Can perform image filtering, morphological operations and other routine operations
5. Can do image segmentation, area counting, geometric measurement and density analysis.
6. Is able to perform data analysis, filtering, statistical analysis and others related to the parameters extracted from the image.
7. Easy of use/development for machine-learning applications.